The present study was to determine the effect of 8-week of the concurrent exercise training on Murf-l and Atrogin-1 Gene Expression of the vastus lateralis muscle in male Wistar rats.
Material and methodsThis study was conducted as an experimental project consisting of four groups of 35 two-month-old male Wistar rats. The animals were randomly divided in 4 groups: (1) endurance training, (2) resistance training, (3) combined training, and (4) control. The animals in the training groups took part in training programs for 8-week. 48h after the last exercise session, the Murf-1 and Atrogin-1 genes of the vastus lateralis muscle were examined through the use of qPCR method.
ResultsThe results obtained from this study revealed that after 8-week of endurance exercise, Murf-1 and Arogin-1 gene expression significantly increased compared to the control group (p=0.04 and p=0.043). In contrast, in the resistance training group, the gene expression of Murf-1 and Atrogin-1 decreased significantly in comparison with the control group (p=0.02 and p=0.04). In addition, the concurrent training group showed no difference in Murf-1 and Atrogin-1 gene expression after 8-week of exercise compared to that of the control group (p=0.43 and p=0.38).
ConclusionsBased on the results of the present research, it can be expressed that resistance training prevents muscular atrophy by decreasing Murf-1 and Atrogin-1 gene expression. Conversely, endurance exercises cause an increase in the expression of these genes, thereby leading to atrophy in the muscles. The results also showed that concurrent exercises do not have a meaningful effect on muscular atrophy.
Skeletal muscles play an influential part in health and illnesses. Having a suitable muscle volume is essential to health and survival. It has a significant role in mobility, morbidity, and mortality. Synthesis and destruction of proteins are part of the natural functioning of metabolism and hemostasis, which enables the muscles to make adjustments according to the stimulator and the imposed load.1 Muscular atrophy is caused by the reduction in synthesizing proteins and probably an increase in protein destruction.2 Muscle mass atrophy includes reduction in the number of muscle fibers (hyperplasia) and reduction in the size of muscle fibers (atrophy).3 Rapid muscle atrophy is one the disabling factors common in the most systemic diseases such as diabetes, cancer, hyperthyroidism, and uremia, and it occurs in certain muscles as a result of immobility or trauma. The different types of muscular atrophy are characterized by a series of biochemical changes.4 The three main proteolytic pathways in the skeletal muscle are: (1) the lysosmal system, (2) the cytosolic Ca2+ activated system, and (3) the ubiquitin proteasome system (UPS). Although all of these pathways are involved in muscle protein breakdown, it occurs through UPS in more than 80 percent of proteolysis during the atrophy of the skeletal muscle. Immobility related atrophy is the most common type of atrophy which occurs due to the illness or the lack of exercise activity for a long time.2,5 Long-term adaptation to physical training is the most likely due to the cumulative effects of each exercise session, which changes the transcription of certain DNA sequences and alters the messenger RNA amount for translation processes.6 Such molecular changes turn into a fixed new level of specialized proteins, resulting in training adaptation.7 Thus, a resistance training program which causes skeletal muscle hypertrophy is accompanied by an increase in the level of the key cellular proteins and protein synthesis molecules. Furthermore, in parallel with these changes, the levels of proteolytic genes, too, are promptly adjusted, and thus, result in the maintenance of or further increase in the muscle mass following a physical exercise.8 The genomic studies designed for identifying the atrophy process indices have identified two genes whose expression increases considerably in this model of muscular atrophy: Murf-1 and Atrogin-1.3 Therefore, the researchers of the present study have proceeded to investigate the genetic factors affecting health.
Murf-1 has been identified as a specific ring finger muscle protein, which links to Titin A168/A169 close to Titin kinase domain in vitro. Murf-1 is found in the myofibrils within the M-Line network region near the catalyst Titin domain as well as inside the cytoplasm in a solved state. Since the members of the Murf-1 family can form heterodimers and may link the transmission pathways of Titin fibers and microtubules, it becomes clear that Murf-1 regulates the dynamics of the microtubules.9 On the other hand, Atrogin-1 gene encodes a member of F-box protein, which is specified by approximately 40 basic amino acids. F-box protein is one of the four complex subunits of ubiquitin ligase protein called SKP1-Cullin-F-box (SCFs), which is activated through the phosphorylation-dependent ssubiquitination.3,10 On the whole, the finding indicates that Murf-1 and Atrogin-1 expressions are valid indices of muscular atrophy due to UPS system.11
In their research, Ribeiro et al. (2017) demonstrated that resistance exercise was effective in increasing Murf-1 and Atrogin-1 of the soleus muscle as a result of aging. Furthermore, they showed that Murf-1 levels decreased in the exercised rats compared to the non-trained rats.12 In their study, Sheybani et al. (2018) came to the conclusion that FOXO3a and Atrogin-1 significantly decreased after physical exercise and had a meaningful increase during non-training period.13 The increase in Murf-1 was probably due to the change in the type of the fibers. Also, there are other findings indicating that age-related changes in rats affected the gene expression in the control group based on gender; i.e., Murf-1 gene expression increased by 52% in the quadriceps femoris muscle of the male rats and decreased by 72% in the female rats during the 15–23 months of age. However, after adaptation to exercise training, Murf-1 expression was not significantly affected by age. Also, mRNA expression of Murf-1 did not change based on the age, gender or physical exercise.14
In their research, Khorramshahi et al. (2016) concluded that performing high intensity interval training (HIIT) causes significant reduction in Atrogin-1 expression in the gastrocnemius muscle. It seems that high intensity interval training can be an effective factor in reducing muscular atrophy through changing Atrogin-1 and miR-23a expression due to improving hyperglycemia.15
Although several researches have been carried out concerning the effect of endurance and resistance training on Murf-1 and Atrogin-1 gene expression, due to the contradiction in the previous researches and different training protocols, the authors proceeded to conduct a research in which they could study the effect of concurrent training besides the effect of endurance and resistance exercise separately. Accordingly, the goal of the present research is to investigate the effect of 8-week of endurance, resistance, and concurrent physical training on the Murf-1 and Atrogin-1 gene expression in the vastus lateralis muscle of male Wistar rats.
Materials and methodsThe present research was carried out as a basic experimental study on 35 two-month-old male Wistar rats with average body weight of 300±50g. The rats were purchased from Posteur institute, a center for breeding laboratory animals, and were kept and controlled in special cages of transparent polycarbonate with the dimensions 54×34×21cm in the laboratory conditions of 22±2°C temperature, 50±5 humidity, and 12:12 light–dark cycle, with free access to water and food pellets. At different stages of the research, based on the rules of the committee of ethics associated with working with laboratory animals according to the protocol of Institutional Animal Care and Use Committee (IACUC), we avoided any kind of abuse or using any kind of unnecessary methods on the animals. This research has been approved by the Medical Committee of the Faculty of Medical Sciences, Islamic Azad University of Tabriz, with the Ethical code number IR.IAU.TABRIZ.REC.1396.007.
To prevent stress and change of physiological conditions, the subjects were kept under the new conditions for one week. After getting accustomed to the new conditions, the rats were randomly assigned to 4 groups: (1) control group (n=8), and (2) endurance, (3) resistance, and (4) concurrent training groups (each consisting of 9 rats). In the second week, the subjects were introduced to the physical training manner. They were taught how to exercise on the smart animal treadmill (walking on the treadmill at the speed of 5 meters per minute and the inclination of 0 degree) and how to go up the ladder (4–5 repetitions without carrying a weight and with a rest period at the top of the ladder). The concurrent training group became familiar with the combined performance of both endurance and resistance activities. Following the familiarity period, the rats were subjected to 8-week of exercise training.
The resistance training in the first week included going up a 120cm ladder while carrying a weight equal to 50% of the subject's body weight, which was attached to each rat's tail with self-adhesive foam tape. In each training session, there were 8 repetitions of going up the ladder with rest periods on the top of the ladder. At the beginning of each week, the subjects were weighed, and with regard to their weights, 10% was added to the weight of the loads.16 The endurance training group followed the training protocol given in Table 1.17
Endurance exercises protocol.
| Variable | Duration | Speed | Slope | Variable | Duration | Speed | Slope |
|---|---|---|---|---|---|---|---|
| Week | Week | ||||||
| Week 1 | 5min. | 5m/min. | 0 | Week 5 | 5min. | 8m/min. | 5 |
| 5min. | 7m/min. | 0 | 10min. | 13m/min. | 5 | ||
| 5min. | 8m/min. | 0 | 10min. | 15m/min. | 5 | ||
| 5min. | 5m/min. | 0 | 30min. | 18m/min. | 5 | ||
| 5min. | 8m/min. | 5 | |||||
| Week 2 | 5min. | 7m/min. | 0 | Week 6 | 5min. | 8m/min. | 10 |
| 10min. | 8m/min. | 0 | 10min. | 13m/min. | 10 | ||
| 30min. | 10m/min. | 0 | 10min. | 15m/min. | 10 | ||
| 5min. | 7m/min. | 0 | 30min. | 18m/min. | 10 | ||
| 5min. | 8m/min. | 10 | |||||
| Week 3 | 5min. | 8m/min. | 0 | Week 7 | 5min. | 8m/min. | 10 |
| 10min. | 10m/min. | 0 | 10min. | 13m/min. | 10 | ||
| 10min. | 13m/min. | 0 | 10min. | 15m/min. | 10 | ||
| 30min. | 15m/min. | 0 | 30min. | 18m/min. | 10 | ||
| 5min. | 8m/min. | 0 | 5min. | 8m/min. | 0 | ||
| Week 4 | 5min. | 8m/min. | 0 | Week 8 | 5min. | 8m/min. | 10 |
| 10min. | 13m/min. | 0 | 10min. | 13m/min. | 10 | ||
| 10min. | 15m/min. | 0 | 10min. | 15m/min. | 10 | ||
| 30min. | 18m/min. | 0 | 30min. | 18m/min. | 10 | ||
| 5min. | 8m/min. | 0 | 5min. | 8m/min. | 0 | ||
The concurrent training group performed both types (endurance and resistance) of exercise. In order to eliminate the acute effect of the training, 48h after the last training session, sampling and biopsy were performed. The researchers attempted to kill the animals under study within the shortest possible time and with the least pain or torture. The rats were anesthetized through peritoneal injection of the mixture of 3–5mg/kg xylazine and 70mg/kg ketamine, and then a large part of their blood (approximately 4–5ml) was collected for examination in other researches. Next, the rats were operated by experienced professionals, and their vastus lateralis muscles were removed and washed in physiological serum. Through the use of a digital scale, 0.001 of the vastus lateralis muscle of the subjects was measured and kept at −70° temperature to examine the gene expression rate of Murf-1 and Atrogin-1 by Real Time-PCR. Concentrations of RNA were determined by measuring absorbance at 260nm. The purity of the RNA was determined by calculating the absorbance ratio at 260 and 280nm and by ethidium bromide staining. Isolated RNA was stored at −70°C until analysis by quantitative polymerase chain reaction (qPCR). For reverse transcription μg of total RNA was typically used in a reaction containing oligo-dT (500μg/ml), 10mM of each dNTP, 5×first-strand buffer, 0.1M dithiothreitol and 200U of reverse transcriptase (SuperScript II, Invitrogen). Reverse transcription was performed at 70°C for 10min followed by incubation at 42°C for 60min and at 95°C for 10min. Primer sets for rat MuRF-1 and Atrogin-1 were designed using Primer Express software v2.0 (Applied Biosystems, Foster City, CA).
After the extraction of RNA from the muscle tissue according to the Kit Protocol made by Termo Company, USA, the cDNA synthesis kit made by Fermentas Company, Canada, was used for cDNA synthesis. In this research, the qPCR comparative method using SYBR Green color was applied in order to determine the expression rate of Murf-1 and Atrogin-1 genes. For quantitative and qualitative comparison of the rate of Murf-1 and Atrogin-1 gene expression, β-actin reference gene was used through TAKARA kit made in Japan. The sequence of the used primers is shown in Table 2.
For each sample, PCR was performed in duplicate in a 25-μl reaction volume of 5–20ng of cDNA, 12.5μl SYBR Green Master Mix (Applied Biosystems) and 100–200nM of each primer. PCR analyses were carried out using the following cycle parameters: 50°C for 2min, 95°C for 10min followed by 40 cycles of 95°C for 15s and 60°C for 1min. Fluorescence was quantified and analyses of amplification plots were performed with the AB 7300 Sequence Detector System (Applied Biosystems). Results were expressed using the comparative cycle threshold (Ct) method as described by the manufacturer.
First, for assessing the normal distribution of the data and the homogeneity of variances, Shapiro–Wilk and Leven's test were applied, respectively. One-way analysis of variance test was used to evaluate differences between control and exercise groups. p-Values<0.05 were considered statistically significant. Statistical analyses were performed by using SPSS program (version 24 for windows) (SPSS Inc. Chicago, IL, USA).
ResultsBased on the statistic F=0.53 and p=0.66 at the significance level of 0.05, the weight of the animals in the four groups of endurance, resistance and concurrent training, and control didn’t have a significant difference prior to the intervention. With regard to Table 3, after two months of endurance, resistance, and concurrent training, there was a meaningful change in the weight of the three training groups. The post hoc test indicated that after the intervention, there was not a significant change in the weight of the control group rats (p=0.33); however, the mean weight of the rats in the endurance, resistance, and concurrent training groups significantly decreased compared to the mean weight before the start of the intervention (p=0.007, p=0.02, and p=0.03, respectively).
Comparison of the weight and gene expression of Murf-1 and Atrogin-1 after 8-week of intervention.
| Variable | Group | Number | Mean | Standard deviation | F | p |
|---|---|---|---|---|---|---|
| Weight (g) | Endurance | 9 | 310.4 | 6.6 | 5.29 | 0.005 |
| Resistance | 9 | 304.2 | 10.8 | |||
| Concurrent | 9 | 316.8 | 7.8 | |||
| Control | 8 | 349.2 | 7.9 | |||
| Muscle weight (mg) | Endurance | 9 | 1.29 | 0.27 | 1.16 | 0.341 |
| Resistance | 9 | 1.13 | 0.18 | |||
| Concurrent | 9 | 1.24 | 0.12 | |||
| Control | 8 | 1.22 | 0.13 | |||
| Murf-1 (ct) gene expression | Endurance | 9 | 24.48 | 0.26 | 17.39 | 0.001 |
| Resistance | 9 | 22.02 | 0.23 | |||
| Concurrent | 9 | 24.04 | 0.28 | |||
| Control | 8 | 23.44 | 0.27 | |||
| Atrogin-1 (ct) gene expression | Endurance | 9 | 24.97 | 0.17 | 13.05 | 0.001 |
| Resistance | 9 | 22.47 | 0.17 | |||
| Concurrent | 9 | 23.11 | 0.47 | |||
| Control | 8 | 23.78 | 0.27 | |||
Significance level equals p<0.05.
Table 3 illustrates the means and standard deviations of the weight and the gene expression of Murf-1and Atrogin-1 of the vastus lateralis muscle of the rats in the endurance, resistance, and concurrent training groups as well as the control group after performing the intervention. It also shows the results of comparing the four groups through the use of one-way analysis of variance test.
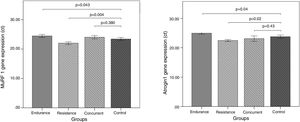
Murf-1 and Atrogin-1 gene expression levels of the vastus lateralis muscle of the male Wistar rats in the endurance, resistance, concurrent training, and control groups, as well as the comparison of Murf-1 and Atrogin-1 gene expression levels between the training groups and the control group are present in Fig. 1.
Murf-1 gene expression levels of the concurrent and endurance training groups increased 2.58% and 4.44% more than the control group, respectively, and Atrogin-1 gene expression levels of the resistance training group decreased 6.04% less than the control group. Atrogin-1 gene expression levels of the concurrent and resistance training groups decreased 2.79% and 5.82% less than the control group, respectively, and Atrogin-1 gene expression levels of the endurance training group increased 4.77% more than the control group.
The results of qPCR indicate that the level of Murf-1 and Atrogin-1 gene expression significantly increased following the endurance training, compared to the control group (p=0.043 and p=0.04). On the contrary, in the three training groups, there was a meaningful reduction in the Murf-1 and Atrogin-1 gene expression levels (p=0.04 and p=0.04). However, after the concurrent training, there was not a significant difference in the level of Murf-1 and Atrogin-1 gene expression in comparison with the control group (p=0.38 and p=0.43).
DiscussionThe present study is among the few research addressing the role of concurrent physical training in the Murf-1 and Atrogin-1 gene expression. The results of the research indicated that performing 8-week of concurrent training did not have any effect on the expression of Murf-1 and Atrogin-1 genes, but 8-week of endurance training caused an increase in the expression of the genes involved in atrophy pathway. The oxidative enzyme regulator in some cell varieties, such as skeletal cells, is PGC1α,18 which acts strongly in oxidative metabolism, and its expression in skeletal muscles induces many of the changes that depend on the endurance activities, namely mitochondrial biogenesis, changes in the type of the muscle fiber, and resistance against atrophy.19 It has been determined that FOXO, the protein that breaks up muscle tissue, and PGC1α cofactor have reciprocal effects.18 FOXO protein is a subdivision of the large family of transcription factors. This family is characterized by DNA-bound demains.20 FOXO3a protein is expressed in skeletal muscles.21 FOXO3 is found in cytosol. Evidence has shown that FOXO3a is the main mediator in the process of atherogenesis, and if it is activated, it is moved from the cytosol to the nucleus and causes the activation of the two proteins destroying muscle tissues, namely Murf-1 and Atrogin-1. FOXO3a can directly activate atrogin-1 and cause atrophy.22 It has been reported that endurance exercises can increase autophagic indices in skeletal muscles.23,24 Other research confirms the results of the present study by showing that running at 60% intensity caused an increase in Atrogin-1 gene expression in the 8-week-old trained rats.25 The reason for the increase in these ligases might be the adaptation response, since it results in facilitating the process of rebuilding as well as the removal of the impaired proteins. Also, Sheybani et al. (2018) reported hypertrophy after training due to the reduction in FOXO3a and Atrogin-1 levels.13 Murf-1 is a ubiquitin ligase exclusive to muscles, which can facilitate the destruction of muscle fibers during atrophy.26 The results of other research show that mRNA levels of Murf-1 and the activity of Proteasomes, as well as the Calpain and Cathepsin levels increase immediately after physical training; however, they return to normal levels after 24h. The level of Murf-1 mRNA, the activity of proteasomes, and abundance of Calpain in the rats that had performed endurance training for eight weeks were studied. Proteasome activity decreased in the rats that had exercised for two weeks. However, the researchers found that the mRNA of the ubiquitin ligases increased after eight weeks of training, accompanied by hypertrophy in the plantaris muscle. The findings of the study indicated that Murf-1 and Atrogin-1 have a role not only in atrophy but in hypertrophy as well.27 The examinations carried out in other research on a number of proteins involved in atrophy showed that Atrogin-1, Murf-1, and FOXO did not change under any of the conditions.28 Murf-1 is considerably affected by the duration of training,27 which could probably be the main reason for the contradictory results of the researches. In one study, endurance training caused an increase in muscle weight and muscle fiber size in diabetic rats. Diabetes induced the increase in Murf-1 expression, and there was a significant decrease in Murf-1 expression in the diabetic group after physical exercise, compared to the control group. These results indicate that physical exercise has an important role in controlling Murf-1 expression in diabetes.29 Furthermore, Khoramshahi et al. (2015) concluded that performing HIIT leads to a significant decrease in Atrogin-1 expression in the gastrocnemius muscle. It seems that HIIT training can be an effective factor in reducing muscle atrophy through improving hyperglycemia and changing Atrogin-1 and miR-23a expression.15
As well, according to the results of the present research, after 8-week of resistance training, the rate of Murf-1 and Atrogin-1 gene expression was decreased. In line with these results, Ribeiro et al. (2017) demonstrated that resistance training is effective in coping with the increase of Murf-1 and Atrogin-1levels in the soleus muscle due to aging and that resistance training decreased the rate of FOXO expression. In addition, Murf-1 levels decreased in the young trained rats in comparison with the young non-trained rats. As a result, by performing resistance exercise, intramyocellular lipid accumulation is reduced, which results in the reduction of Peroxisome proliferator-activated receptor γ gene expression; in other words, the glucose hemostasis regulators become limited. This response is related to the catabolic reduction of FOXO, Atrogin-1, and Murf-1 axis, as well as to the anabolic increase of the IGF-1, mTOR, and MyoD axis.12 In another study, by performing resistance exercise on the femoral muscle, the rate of FOXO expression was higher in the control group than any of the other groups. Atrogin-1 expression in the control and training groups was higher than that in the other groups. The expression of FOXO did not show a significant difference in the soleus muscle, but the expression of Atrogin-1 and Murf-1 decreased in the training group, compared to the control group.30 Furthermore, Zanchi et al. (2009) came to the conclusion in their research that resistance training causes reduction in Murf-1 and Atrogin-1 gene expression (41.64% and 61.19%, respectively)8; however, another group of researchers, after conducting resistance exercises with a different protocol, reached the conclusion that resistance training does not have any impact on the expression of these genes.14 In conclusion, with regard to the results of the present research, after conducting 8-week of endurance, resistance, and concurrent exercise training with upper-medium intensity, it can be concluded that endurance exercise trainings increase the expression of Murf-1 and Atrogin-1 genes. On the contrary, performing resistance exercise training reduces these two genes. Also, it can be stated that performing 8-week of concurrent physical training does not have any impact on the expression of Murf-1 and Atrogin-1 genes. Thus, resistance exercise trainings prevent muscle atrophy by reducing Murf-1 and Atrogin-1 gene expression, but endurance training increases the expression of these genes, which in turn leads to atrophy in the muscles. Furthermore, one of the functional results of this research is the weight loss of the subjects. One of the main concerns of today's human societies is losing weight, which can be achieved by taking into account the results obtained from this study and performing endurance, resistance, and especially concurrent exercise training. With regard to the limitations of the present research, such as lack of measurement of morphological changes and evaluation of the expression of the other proteins involved in atrophy, making definitive statements regarding the effect of different exercise trainings on the vastus lateralis muscle atrophy requires further research and study.
Statement of financial supportThis research is not funded by a specific project grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of interestThis research authors declare no conflict of interest.
The study was designed by FZ and AN; data were collected and analyzed by YM and FZ; manuscript preparation and data interpretation were undertaken by FZ, YM and AN. All authors approved the final version of the paper.